FEBRUARY 2022
Kyra's Blog
Kyra Bankhead, undergraduate student
1 February 2022
After editing the methods section of draft with Alejandro I realized I messed up measuring overdispersion in the response variable at the waterfront site. I used two different methods to check for overdispersion at the waterfront and both came back with overdispersion detected. I tried to reanalyze Cambell’s 2021 article on “The consequences of checking for zero-inflation and overdispersion in the analysis of count data,” and found that both the waterfront and marina follow a negative binomial distribution as this family treats for both zero inflation and overdispersion. However, when a negative binomial distribution is used for the waterfront, the results do not make sense. When the number of harbor seals hauled-out are graphed with ambient noise level, there is clearly no correlation as the data points are widely dispersed (see below). So there is something essential that I am still missing and I cannot continue on to the results until I figure it out.
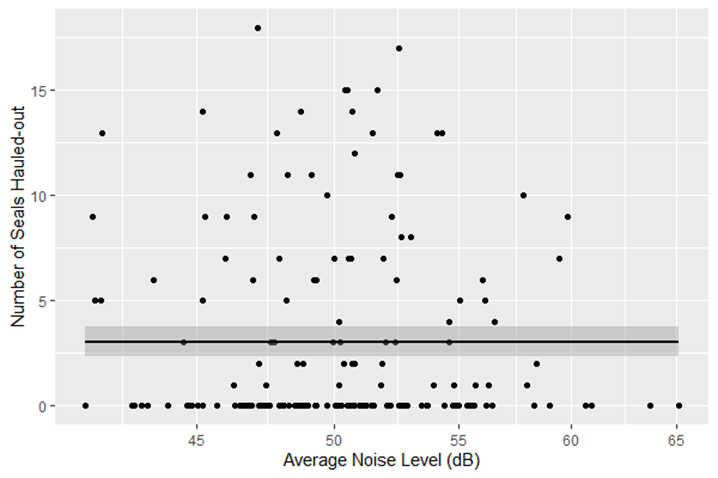
Scatter plot of number of seals hauled-out versus the average noise level in decibels during a 30-min observation period at the Bellingham Waterfront. The gray area around the regression line shows 95% confidence intervals.
Until then,
Kyra Bankhead
Madison's Blog
Madison Gard, undergraduate student
1 February 2022
First blog ever! Big moment! January was a good month of getting back into a routine with school, work, and research along with some chaotic shifts from remote work to on-campus and back again (thank you, pandemic). A silver lining of remote work was spending more time with my new cat, Otis 🙂

Ottis the cat.
My research focus has been getting everything running again for sample processing, as well as orienting myself to begin drafting the manuscript on q-PCR methods validation for sex determination in Steller and California sea lions. Zoë Lewis has been an awesome mentor and resource throughout this process. We started out ahead of the game because we had prepped half of the aquarium samples by portioning and drying them before winter break. This let us hit the ground running once winter quarter began. In January, I finished portioning and drying the second half of aquarium samples which are now ready to be processed using the Qiagen Fast DNA Stool Protocol kit. We also processed the first batch of samples using the Qiagen protocol, and after checking for DNA using the Qubit fluorometer, we ran our first plate through the qPCR instrument!
Outside of the lab, I’ve been reading published literature and annotating with helpful notes for once I’m ready to start writing. I also created an outlined schedule for the goals I’d like to meet each week to keep me on track. My ultimate goal is to have a first draft of the manuscript ready by the end of spring quarter. I’m stoked for this opportunity and ready to work!
Thank you!
Maddie Gard
Holland's Blog
Holland Conwell, undergraduate student
1 February 2022
With the surge in covid cases and bouncing between virtual and in-person classes, it’s certainly turned out to be an unusual start to the quarter! That being said, an unanticipated silver lining of beginning the quarter online turned out to be having more free time to work on this project. Since last month, I’ve resolved my troubles in adding and sorting the overall sex/diet dataset by orders. I was originally having some difficulties with formatting, but I resolved this by keeping the diet dataset in Excel and simply using a previous code to combine all the other information into this spreadsheet. This helped me avoid all the strange formatting issues that I was encountering in RStudio. Regarding the sexing data, I merged this with the sample information from scratch, using the updated sexing results from the plates that were rerun. Following this merge, I then combined this sheet with the diet data, attributed orders to species in the dataset, and added in sex/diet data from Madelyn Voelker’s previous study in the lab.
Now that I have all this information more or less in the same place, I decided to revisit the related Schwarz 2018 paper for a loose roadmap of statistical analyses. I put together my own outline of these methods and refreshed my memory on how to conduct a few of these. Preceding the generalized linear models and PERMANOVA analyses, it certainly seems that I should start with analyzing seal sex ratios at haul out sites. I began putting together a pivot table in Excel that would churn out these ratios, and even though I got results, I realized that until the dataset is condensed to a wide format, I won’t be able to get accurate ratios. Currently, I’m working on this step and looking ahead to the analyses to come in the next month!
Kathleen's Blog
Kathleen McKeegan, graduate student
1 February 2022
Winter quarter started off a little rocky, with switches from virtual learning to in-person and then back to virtual and now finally in-person again. Luckily, my project is at a stage where I can easily work from home or from the office/lab. I feel very fortunate to be able to continue working on my master’s project throughout these crazy times.
This past month, Kate and I continued to work on processing the 2021 photographic data. The protocol for completely processing these photos is incredibly time consuming. I began getting concerned about our timeline, worried that we would be unable to finish processing this data in time for my scheduled spring graduation. So with that in mind, she and I decided to alter the protocol slightly. Instead of selecting, cropping, IDing, and naming each photo of each seal per minute of each observation, we decided to focus on determining which seals were there on the day and which seals have a fish. The more detailed minute-by-minute work will be completed later with help from our team of research assistants. This switch in protocol will allow us to gather the necessary information from 2021 much quicker, and therefore I can incorporate this into my data analysis on schedule.
Given this switch, my methods for analysis have changed slightly. I am going to analyze the TAST-on year (2020) at a finer scale, looking at the days in a minute-by-minute window to allow for comparison between TAST on and TAST off days in 2020. When comparing across the years (2019-before, 2020-during, 2021-after), I will look at the data in a broader view, comparing which seals are there on which days and looking at who has a fish. So far, I have started the fine-scale 2020 analysis, summarizing the presence/absence of IDs across the run-season. For example, I found that ID 0173 was present the most between Oct-Dec in 2020, showing up for 753 ‘occurrences’, or minutes observed. I am still working on coding my analysis in R, but I will have most (if not all) of my results completed over the next month.
Other than analysis, our team is conducting 1-2 observations per week to get year-long data on the seals at the creek. As with previous years, very few (if any) seals are present at this time of year. So far, we’ve only seen ID 0012, the seal that, for whatever reason, loves to hang out in the creek during the off-season. I am also working on more thesis drafts, currently focusing on my methods and results sections. Further, I am very excited to be giving a presentation to the Bellingham Technical College Fisheries students at the Whatcom Creek Hatchery in a few weeks! I am looking forward to hearing their opinion on the pinniped-fisheries conflict here in the Pacific Northwest. Shortly after that, I will be presenting my research at the Whatcom Marine Research Symposium. Suffice it to say, I will be very busy this month! I am feeling confident but a little overwhelmed by my to-do list. That said, this is the exciting part of the project! I will have more results to share next month and a couple presentations under my belt! We’re (hopefully) in the homestretch!
Zoë's Blog
Zoë Lewis, graduate student
1 February 2022
As winter quarter keeps moving forward, I’ve tried my best to stay on top of new drafts of thesis writing and data analysis. After writing the general introduction, background, and methods of my thesis I’m starting to feel like my thesis is taking shape. Although there are still many more steps to go, I’m excited to start to see these results take shape. Despite some serious data delays… I’m excited to start digging into data analysis this next month.
Unfortunately, I am still waiting on both my hard parts and DNA metabarcoding results, as both contractors have had a few small hiccups in the processing. I’m hoping that by the end of this week, everything will be cleared up and I will have a more complete picture of my data set. Given COVID delays, I’m not surprised that everything has taken a bit longer than expected. Luckily, in the meantime I’ve been reviewing papers and statistics to help me outline my analysis methods. I always find it a bit challenging to create code to run statistic tests without data, but I find that I better understand this analysis when I am forced to think about the process conceptually first.
On the bright side, the extracts for the Steller sea lion and California sea lions of known sex are complete and the preliminary results look promising for the sexing project! After going through previous methods papers and showing Maddie the initial techniques, she is taking charge of the project. While I’m still around for her to ask questions, Maddie is eager to make this project her own, which is very impressive! Molecular experiments like this take planning, attention to detail, focus, time, and money… so I’m excited to see how far she has come since starting in the lab.
Hopefully, by the next blog I’ll have some interesting data news to share! Until then…